MOGDx Main Functions and Classes
We provide a description of the main functions and classes used to train a MOGDx model.
Graph Neural Network with Multi Modal Encoder (GNN-MME)
The GNN-MME is the main component in the architecture of MOGDx. It consists of a Multi-Modal Encoder (MME) to reduce the dimension of the input modalities and decodes all modalities to a shared latent space.
Along with the MME there is a Graph Neural Network (GNN). Currently there are two GNN’s implemented ; Graph Convolutional Network (GCN) for applications in the transductive setting and GraphSage for applications in the inductive setting. Both algorithms are implemented using the Deep Graph Library (DGL).

Multi Modal Encoder
The MME takes any number of modalities and compresses them to an arbitrary dimension, found through hyperparameter searching. The architecture for the MME has been established in similar research in [Yang et al., 2021] and [Xu et al., 2019].
Graph Convolutional Network (GCN)
GCN, developed by [Kipf and Welling, 2017], was implemented from the DGL. For a tutorial on the use of GCN’s we refer you to the DGL Tutorial Page
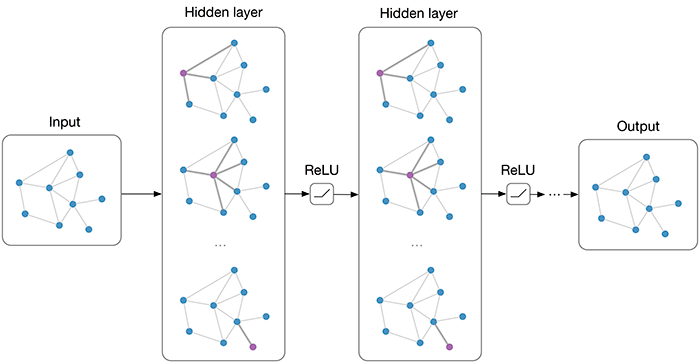
GraphSage
GraphSage, developed by [Hamilton et al., 2017], was implemented from the DGL. For a tutorial on the use of GraphSage we refer you to the DGL Tutorial Page

Training
Functions used to train and evaluate the MOGDx model. The training implemented follows that outlined by [Hamilton, 2020].
The list of functions are :
train
evaluate
confusion_matrix
AUROC
Utility
Utility functions used to parse input data, load networks from csv or perform utility tasks.
The list of functions are :
data_parsing_python
data_parsing_R
get_gpu_memory
indices_removal_adjust
network_from_csv
Citations
Will Hamilton, Zhitao Ying, and Jure Leskovec. Inductive Representation Learning on Large Graphs. In Advances in Neural Information Processing Systems, volume 30. Curran Associates, Inc., 2017. URL: https://proceedings.neurips.cc/paper_files/paper/2017/hash/5dd9db5e033da9c6fb5ba83c7a7ebea9-Abstract.html (visited on 2024-02-12).
William L. Hamilton. Graph Representation Learning. Synthesis Lectures on Artificial Intelligence and Machine Learning. Springer International Publishing, Cham, 2020. ISBN 978-3-031-00460-5 978-3-031-01588-5. URL: https://link.springer.com/10.1007/978-3-031-01588-5 (visited on 2024-03-13), doi:10.1007/978-3-031-01588-5.
Thomas N. Kipf and Max Welling. Semi-Supervised Classification with Graph Convolutional Networks. arXiv, February 2017. arXiv:1609.02907 [cs, stat]. URL: http://arxiv.org/abs/1609.02907 (visited on 2022-09-26).
Jing Xu, Peng Wu, Yuehui Chen, Qingfang Meng, Hussain Dawood, and Hassan Dawood. A hierarchical integration deep flexible neural forest framework for cancer subtype classification by integrating multi-omics data. BMC Bioinformatics, 20(1):527, October 2019. URL: https://doi.org/10.1186/s12859-019-3116-7 (visited on 2024-03-15), doi:10.1186/s12859-019-3116-7.
Hai Yang, Rui Chen, Dongdong Li, and Zhe Wang. Subtype-GAN: a deep learning approach for integrative cancer subtyping of multi-omics data. Bioinformatics, 37(16):2231–2237, August 2021. URL: https://doi.org/10.1093/bioinformatics/btab109 (visited on 2024-03-15), doi:10.1093/bioinformatics/btab109.