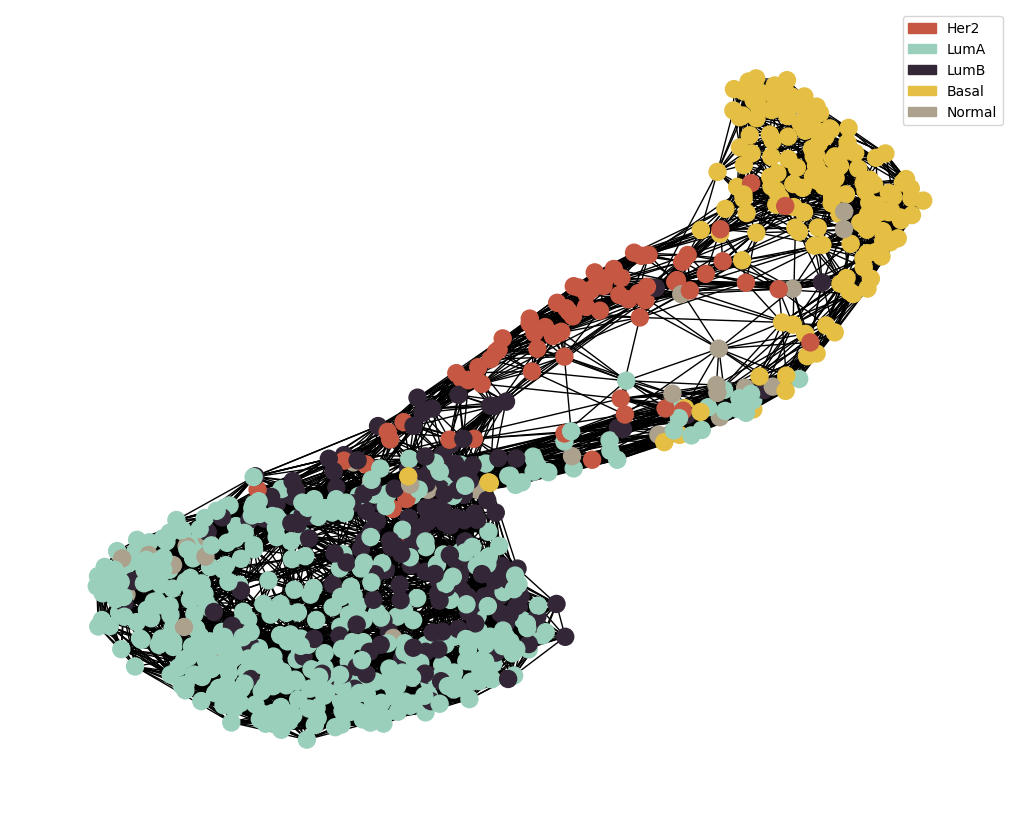
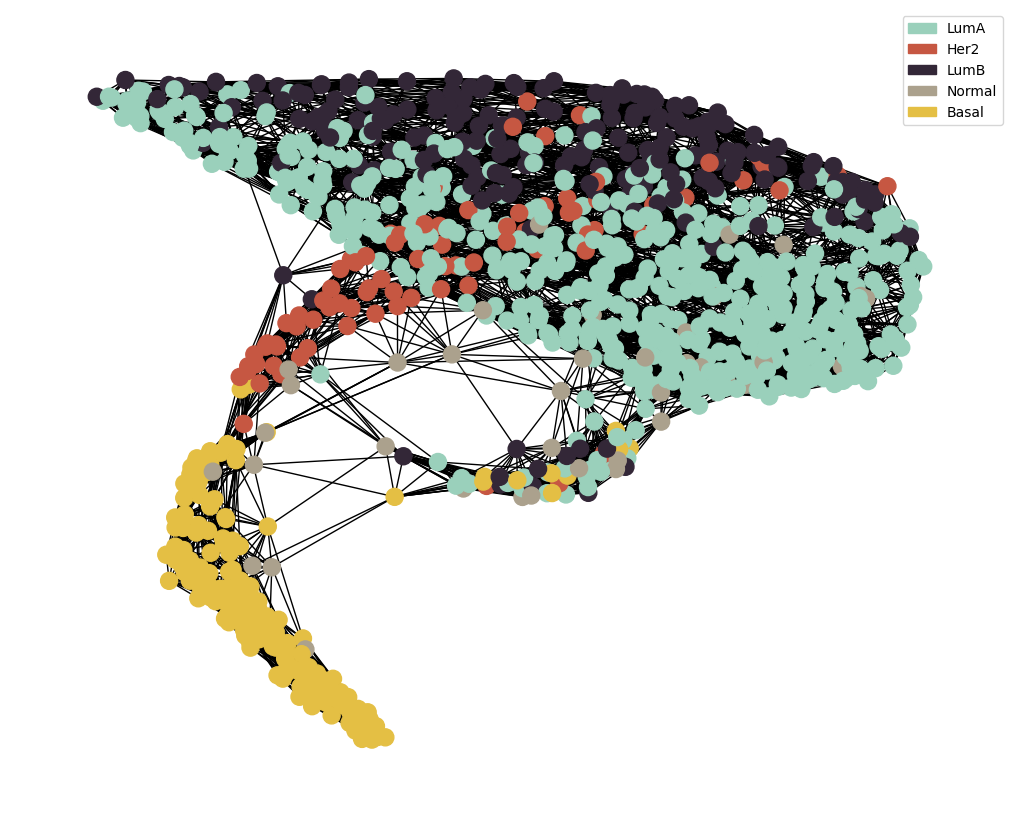
MOGDx Network Generation
This is the computationally most expensive architecture, but requires the least amount of knowledge.
The processing step occurs within each cross validation split such that similarity features are only identified within each split.
import pandas as pd
import numpy as np
import os
import sys
sys.path.insert(0 , './../')
from MAIN.utils import *
from MAIN.train import *
import MAIN.preprocess_functions
from MAIN.GNN_MME import GCN_MME , GSage_MME , GAT_MME
import torch
import torch.nn.functional as F
import dgl
from dgl.dataloading import MultiLayerFullNeighborSampler
import matplotlib.pyplot as plt
from palettable import wesanderson
from sklearn.model_selection import StratifiedKFold , train_test_split
import networkx as nx
from datetime import datetime
import joblib
import warnings
import gc
import copy
warnings.filterwarnings("ignore")
print("Finished Library Import \n")
Finished Library Import
data_input = './../../data/TCGA/BRCA/raw/'
snf_net = 'RPPA_mRNA_graph.graphml'
index_col = 'patient'
target = 'paper_BRCA_Subtype_PAM50'
device = torch.device('cpu' if False else 'cuda') # Get GPU device name, else use CPU
print("Using %s device" % device)
get_gpu_memory()
datModalities , meta = data_parsing(data_input , ['RPPA' , 'mRNA'] , target , index_col , PROCESSED=False)
meta = meta.loc[sorted(meta.index)]
skf = StratifiedKFold(n_splits=2 , shuffle=True)
print(skf)
data_to_save = {}
output_metrics = []
test_logits = []
test_labels = []
for i, (train_index, test_index) in enumerate(skf.split(meta.index, meta)) :
h = {}
MME_input_shapes = []
train_index , val_index = train_test_split(
train_index, train_size=0.8, test_size=None, stratify=meta.iloc[train_index]
)
train_idx = list(meta.iloc[train_index].index)
val_idx = list(meta.iloc[val_index].index)
test_idx = list(meta.iloc[test_index].index)
all_graphs = {}
data_to_save[i] = {}
for mod in datModalities :
count_mtx = datModalities[mod]
datMeta = meta.loc[count_mtx.index]
datMeta = datMeta.loc[sorted(datMeta.index)]
count_mtx = count_mtx.loc[datMeta.index]
train_index_mod = list(set(train_idx) & set(datMeta.index))
val_index_mod = list(set(val_idx) & set(datMeta.index))
if mod in ['mRNA' , 'miRNA'] :
pipeline = 'DESeq'
if mod == 'mRNA' :
gene_exp = True
else :
gene_exp = False
else :
pipeline = 'LogReg'
if pipeline == 'DESeq' :
print('Performing Differential Gene Expression for Feature Selection')
count_mtx , datMeta = MAIN.preprocess_functions.data_preprocess(count_mtx.astype(int).astype(np.float32),
datMeta , gene_exp = True)
train_index_mod = list(set(train_index_mod) & set(count_mtx.index))
dds, vsd, top_genes = MAIN.preprocess_functions.DESEQ(count_mtx , datMeta , target ,
n_genes=500,train_index=train_index_mod)
data_to_save[i][f'{mod}_extracted_feats'] = list(set(top_genes))
datExpr = pd.DataFrame(data=vsd , index=count_mtx.index , columns=count_mtx.columns)
elif pipeline == 'LogReg' :
print('Performing Logistic Regression for Feature Selection')
n_genes = count_mtx.shape[1]
datExpr = count_mtx.loc[: , (count_mtx != 0).any(axis=0)] # remove any genes with all 0 expression
train_index_mod = list(set(train_index_mod) & set(datExpr.index))
val_index_mod = list(set(val_index_mod) & set(datExpr.index))
extracted_feats , model = MAIN.preprocess_functions.elastic_net(datExpr , datMeta ,
train_index=train_index_mod ,
val_index=val_index_mod ,
l1_ratio = 1 , num_epochs=1000)
data_to_save[i][f'{mod}_extracted_feats'] = list(set(extracted_feats))
data_to_save[i][f'{mod}_model'] = {'model' : model}
del model
if mod in [] :
method = 'bicorr'
elif mod in [ 'mRNA' , 'RPPA' , 'miRNA' , 'CNV' , 'DNAm'] :
method = 'pearson'
elif mod in [] :
method = 'euclidean'
datExpr = datExpr.loc[datMeta.index]
if len(data_to_save[i][f'{mod}_extracted_feats']) > 0 :
G = MAIN.preprocess_functions.knn_graph_generation(datExpr , datMeta , method=method ,
extracted_feats=data_to_save[i][f'{mod}_extracted_feats'],
node_size =150 , knn = 15 )
else :
G = MAIN.preprocess_functions.knn_graph_generation(datExpr , datMeta , method=method ,
extracted_feats=None, node_size =150 , knn = 15 )
all_graphs[mod] = G
h[mod] = datExpr
MME_input_shapes.append(datExpr.shape[1])
all_idx = list(set().union(*[list(h[mod].index) for mod in h]))
train_index = indices_removal_adjust(train_index , meta.reset_index()[index_col] , all_idx)
val_index = indices_removal_adjust(val_index , meta.reset_index()[index_col] , all_idx)
test_index = indices_removal_adjust(test_index , meta.reset_index()[index_col] , all_idx)
train_index = np.concatenate((train_index , val_index))
gc.collect()
torch.cuda.empty_cache()
print('Clearing gpu memory')
get_gpu_memory()
if len(all_graphs) > 1 :
full_graphs = []
for mod , graph in all_graphs.items() :
full_graph = pd.DataFrame(data = np.zeros((len(all_idx) , len(all_idx))) , index=all_idx , columns=all_idx)
graph = nx.to_pandas_adjacency(graph)
full_graph.loc[graph.index , graph.index] = graph.values
full_graphs.append(full_graph)
adj_snf = MAIN.preprocess_functions.SNF(full_graphs)
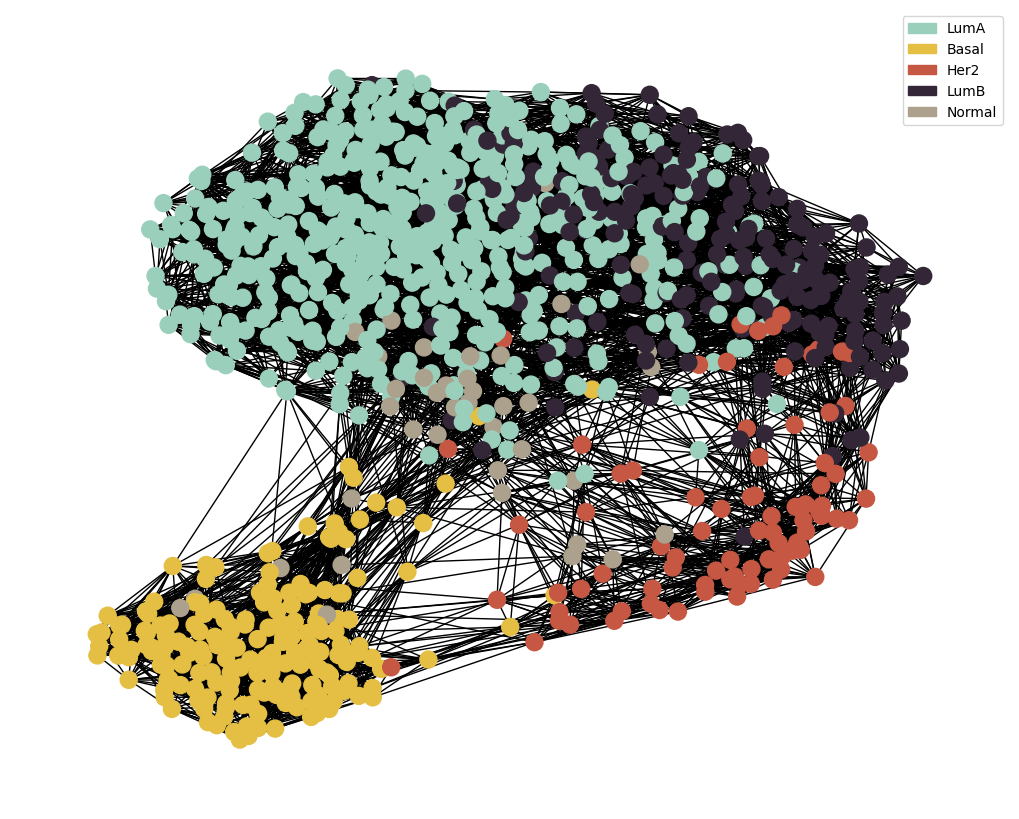
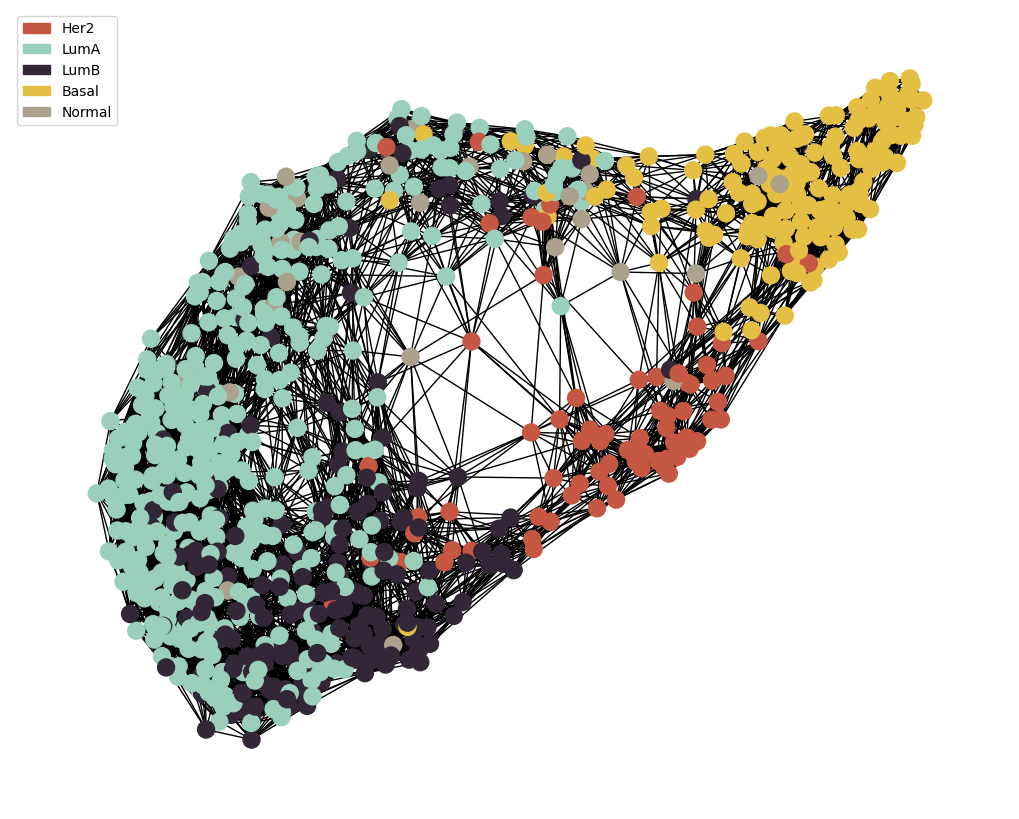
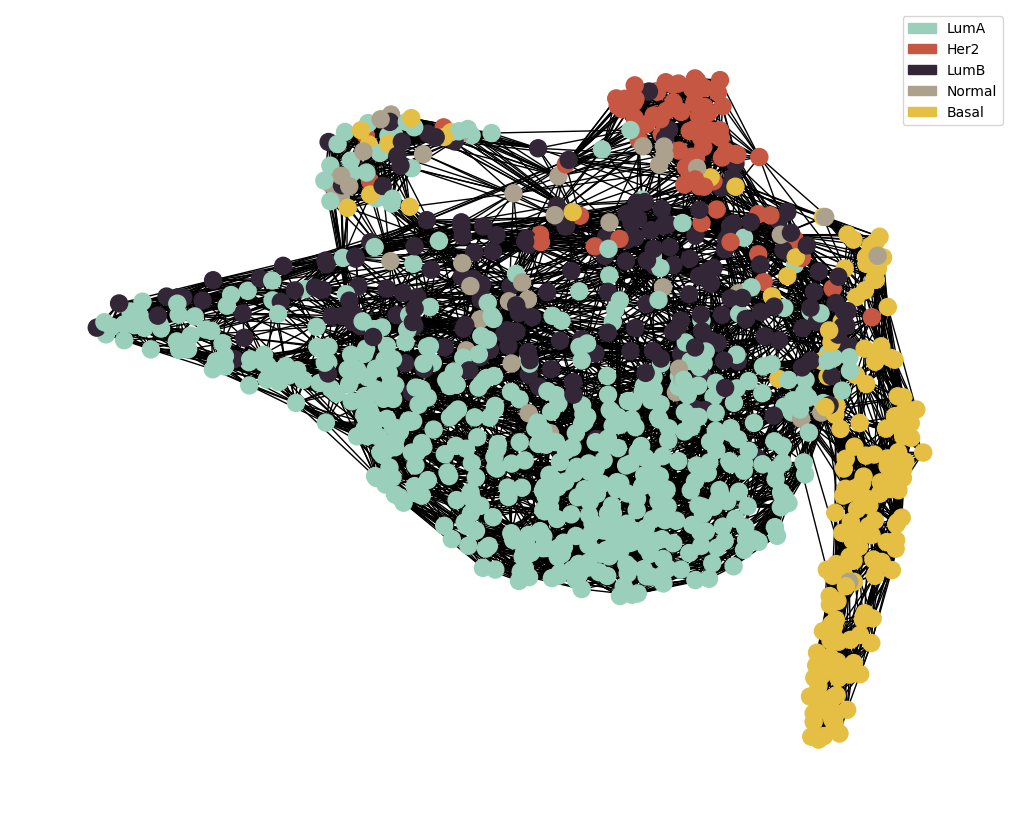
node_colour = meta.loc[adj_snf.index].astype('category').cat.set_categories(wesanderson.FantasticFox2_5.hex_colors , rename=True)
g = MAIN.preprocess_functions.plot_knn_network(adj_snf , 15 , meta.loc[adj_snf.index] ,
node_colours=node_colour , node_size=150)
else :
g = list(all_graphs.values())[0]
label = F.one_hot(torch.Tensor(list(meta.loc[sorted(all_idx)].astype('category').cat.codes)).to(torch.int64))
h = reduce(merge_dfs , list(h.values()))
h = h.loc[sorted(h.index)]
g = dgl.from_networkx(g , node_attrs=['idx' , 'label'])
g.ndata['feat'] = torch.Tensor(h.to_numpy())
g.ndata['label'] = label
model = GCN_MME(MME_input_shapes , [16 , 16] , 64 , [32], len(meta.unique())).to(device)
print(model)
print(g)
g = g.to(device)
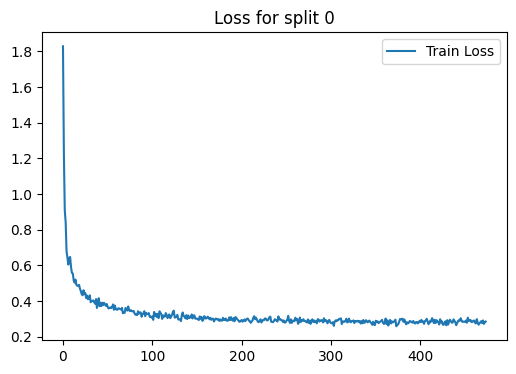
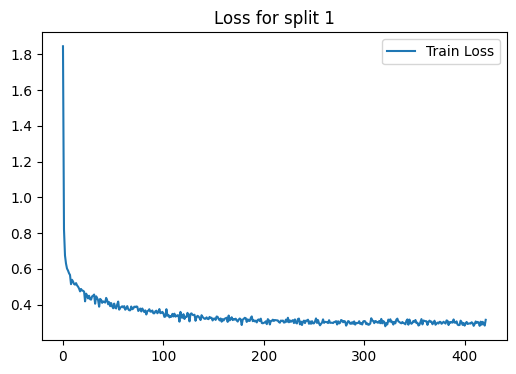
loss_plot = train(g, train_index, device , model , label , 2000 , 1e-3 , 100)
plt.title(f'Loss for split {i}')
plt.show()
plt.clf()
sampler = NeighborSampler(
[15 for i in range(len(model.gnnlayers))], # fanout for each layer
prefetch_node_feats=['feat'],
prefetch_labels=['label'],
)
test_dataloader = DataLoader(
g,
torch.Tensor(test_index).to(torch.int64).to(device),
sampler,
device=device,
batch_size=1024,
shuffle=True,
drop_last=False,
num_workers=0,
use_uva=False,
)
test_output_metrics = evaluate(model , g, test_dataloader)
print(
"Fold : {:01d} | Test Accuracy = {:.4f} | F1 = {:.4f} ".format(
i+1 , test_output_metrics[1] , test_output_metrics[2] )
)
test_logits.extend(test_output_metrics[-2])
test_labels.extend(test_output_metrics[-1])
output_metrics.append(test_output_metrics)
if i == 0 :
best_model = model
best_idx = i
elif output_metrics[best_idx][1] < test_output_metrics[1] :
best_model = model
best_idx = i
get_gpu_memory()
del model
gc.collect()
torch.cuda.empty_cache()
print('Clearing gpu memory')
get_gpu_memory()
test_logits = torch.stack(test_logits)
test_labels = torch.stack(test_labels)
Using cuda device
Total = 42.4Gb Reserved = 0.0Gb Allocated = 0.0Gb
StratifiedKFold(n_splits=2, random_state=None, shuffle=True)
Performing Logistic Regression for Feature Selection
Loss : 0.6042: 100%|██████████| 1000/1000 [00:02<00:00, 494.66epoch/s]
Model score : 0.877
Performing Differential Gene Expression for Feature Selection
Keeping 29995 genes
Removed 30665 genes
Keeping 1047 Samples
Removed 36 Samples
Fitting size factors...
Using None as control genes, passed at DeseqDataSet initialization
... done in 1.23 seconds.
Fitting dispersions...
... done in 10.47 seconds.
Fitting dispersion trend curve...
... done in 0.76 seconds.
Fitting MAP dispersions...
... done in 10.90 seconds.
Fitting LFCs...
... done in 7.12 seconds.
Calculating cook's distance...
... done in 1.82 seconds.
Replacing 2948 outlier genes.
Fitting dispersions...
... done in 1.11 seconds.
Fitting MAP dispersions...
... done in 1.02 seconds.
Fitting LFCs...
... done in 1.02 seconds.
Performing contrastive analysis for LumA vs. Her2
Running Wald tests...
... done in 2.55 seconds.
Log2 fold change & Wald test p-value: paper_BRCA_Subtype_PAM50 LumA vs Her2
baseMean log2FoldChange lfcSE stat \
ENSG00000000003.15 3087.392130 -0.045843 0.109944 -0.416972
ENSG00000000005.6 72.999641 2.364056 0.293465 8.055675
ENSG00000000419.13 2386.496112 -0.604066 0.062602 -9.649254
ENSG00000000457.14 1596.783158 -0.062327 0.063247 -0.985452
ENSG00000000460.17 715.140714 -0.435593 0.080773 -5.392793
... ... ... ... ...
ENSG00000288658.1 22.532026 -0.955682 0.240269 -3.977559
ENSG00000288663.1 29.867524 0.627702 0.099567 6.304313
ENSG00000288670.1 422.158728 -0.075705 0.079346 -0.954105
ENSG00000288674.1 8.197550 0.337858 0.120301 2.808436
ENSG00000288675.1 32.370671 -0.912699 0.116411 -7.840287
pvalue padj
ENSG00000000003.15 6.766989e-01 7.320512e-01
ENSG00000000005.6 7.904166e-16 9.221527e-15
ENSG00000000419.13 4.951465e-22 1.092053e-20
ENSG00000000457.14 3.244019e-01 3.944238e-01
ENSG00000000460.17 6.937103e-08 3.022199e-07
... ... ...
ENSG00000288658.1 6.962648e-05 1.878268e-04
ENSG00000288663.1 2.894748e-10 1.756229e-09
ENSG00000288670.1 3.400305e-01 4.104478e-01
ENSG00000288674.1 4.978277e-03 9.501362e-03
ENSG00000288675.1 4.495165e-15 4.869356e-14
[29995 rows x 6 columns]
Performing contrastive analysis for LumA vs. LumB
Running Wald tests...
... done in 2.71 seconds.
Log2 fold change & Wald test p-value: paper_BRCA_Subtype_PAM50 LumA vs LumB
baseMean log2FoldChange lfcSE stat \
ENSG00000000003.15 3087.392130 0.510947 0.075755 6.744707
ENSG00000000005.6 72.999641 0.680790 0.200628 3.393294
ENSG00000000419.13 2386.496112 -0.496193 0.043131 -11.504238
ENSG00000000457.14 1596.783158 0.070789 0.043570 1.624718
ENSG00000000460.17 715.140714 -0.607113 0.055622 -10.914917
... ... ... ... ...
ENSG00000288658.1 22.532026 -0.808325 0.165651 -4.879702
ENSG00000288663.1 29.867524 -0.053662 0.066356 -0.808701
ENSG00000288670.1 422.158728 -0.228263 0.054607 -4.180137
ENSG00000288674.1 8.197550 0.269729 0.080393 3.355135
ENSG00000288675.1 32.370671 0.324366 0.081632 3.973538
pvalue padj
ENSG00000000003.15 1.533361e-11 1.663405e-10
ENSG00000000005.6 6.905757e-04 1.840247e-03
ENSG00000000419.13 1.255933e-30 1.034937e-28
ENSG00000000457.14 1.042227e-01 1.570698e-01
ENSG00000000460.17 9.781815e-28 6.378381e-26
... ... ...
ENSG00000288658.1 1.062460e-06 5.130976e-06
ENSG00000288663.1 4.186870e-01 5.071215e-01
ENSG00000288670.1 2.913337e-05 1.041544e-04
ENSG00000288674.1 7.932621e-04 2.084624e-03
ENSG00000288675.1 7.081273e-05 2.341559e-04
[29995 rows x 6 columns]
Performing contrastive analysis for LumA vs. Normal
Running Wald tests...
... done in 2.58 seconds.
Log2 fold change & Wald test p-value: paper_BRCA_Subtype_PAM50 LumA vs Normal
baseMean log2FoldChange lfcSE stat \
ENSG00000000003.15 3087.392130 -0.149391 0.151267 -0.987602
ENSG00000000005.6 72.999641 -0.070272 0.400423 -0.175495
ENSG00000000419.13 2386.496112 0.052635 0.086204 0.610591
ENSG00000000457.14 1596.783158 0.460586 0.087118 5.286911
ENSG00000000460.17 715.140714 0.442141 0.111457 3.966908
... ... ... ... ...
ENSG00000288658.1 22.532026 -0.735339 0.331180 -2.220363
ENSG00000288663.1 29.867524 0.106228 0.134479 0.789924
ENSG00000288670.1 422.158728 0.475525 0.109506 4.342477
ENSG00000288674.1 8.197550 -0.105403 0.159991 -0.658810
ENSG00000288675.1 32.370671 -0.340401 0.161826 -2.103501
pvalue padj
ENSG00000000003.15 3.233477e-01 0.443638
ENSG00000000005.6 8.606909e-01 0.906762
ENSG00000000419.13 5.414706e-01 0.651681
ENSG00000000457.14 1.243991e-07 0.000002
ENSG00000000460.17 7.281114e-05 0.000401
... ... ...
ENSG00000288658.1 2.639415e-02 0.059055
ENSG00000288663.1 4.295720e-01 0.548929
ENSG00000288670.1 1.408855e-05 0.000097
ENSG00000288674.1 5.100176e-01 0.624223
ENSG00000288675.1 3.542197e-02 0.075375
[29995 rows x 6 columns]
Performing contrastive analysis for LumA vs. Basal
Running Wald tests...
... done in 3.07 seconds.
Log2 fold change & Wald test p-value: paper_BRCA_Subtype_PAM50 LumA vs Basal
baseMean log2FoldChange lfcSE stat \
ENSG00000000003.15 3087.392130 -0.445231 0.080360 -5.540469
ENSG00000000005.6 72.999641 0.121811 0.212744 0.572570
ENSG00000000419.13 2386.496112 -0.502644 0.045765 -10.983180
ENSG00000000457.14 1596.783158 0.450413 0.046261 9.736320
ENSG00000000460.17 715.140714 -1.016645 0.058992 -17.233544
... ... ... ... ...
ENSG00000288658.1 22.532026 -1.948089 0.174996 -11.132162
ENSG00000288663.1 29.867524 0.589213 0.072151 8.166402
ENSG00000288670.1 422.158728 0.042231 0.058005 0.728061
ENSG00000288674.1 8.197550 0.081888 0.085157 0.961606
ENSG00000288675.1 32.370671 -0.328268 0.085826 -3.824821
pvalue padj
ENSG00000000003.15 3.016618e-08 5.818872e-08
ENSG00000000005.6 5.669360e-01 6.009346e-01
ENSG00000000419.13 4.604242e-28 2.082392e-27
ENSG00000000457.14 2.110588e-22 7.768694e-22
ENSG00000000460.17 1.487504e-66 1.983009e-65
... ... ...
ENSG00000288658.1 8.748769e-29 4.064106e-28
ENSG00000288663.1 3.177226e-16 9.097937e-16
ENSG00000288670.1 4.665759e-01 5.029268e-01
ENSG00000288674.1 3.362475e-01 3.704861e-01
ENSG00000288675.1 1.308669e-04 2.016518e-04
[29995 rows x 6 columns]
Performing contrastive analysis for Her2 vs. LumB
Running Wald tests...
... done in 2.57 seconds.
Log2 fold change & Wald test p-value: paper_BRCA_Subtype_PAM50 Her2 vs LumB
baseMean log2FoldChange lfcSE stat \
ENSG00000000003.15 3087.392130 0.556791 0.121299 4.590230
ENSG00000000005.6 72.999641 -1.683266 0.323369 -5.205406
ENSG00000000419.13 2386.496112 0.107873 0.069059 1.562059
ENSG00000000457.14 1596.783158 0.133116 0.069775 1.907792
ENSG00000000460.17 715.140714 -0.171520 0.089082 -1.925423
... ... ... ... ...
ENSG00000288658.1 22.532026 0.147357 0.264721 0.556650
ENSG00000288663.1 29.867524 -0.681364 0.109132 -6.243507
ENSG00000288670.1 422.158728 -0.152558 0.087504 -1.743435
ENSG00000288674.1 8.197550 -0.068130 0.132312 -0.514919
ENSG00000288675.1 32.370671 1.237065 0.128962 9.592483
pvalue padj
ENSG00000000003.15 4.427577e-06 2.483732e-05
ENSG00000000005.6 1.935725e-07 1.438248e-06
ENSG00000000419.13 1.182741e-01 1.871212e-01
ENSG00000000457.14 5.641806e-02 1.010847e-01
ENSG00000000460.17 5.417652e-02 9.782234e-02
... ... ...
ENSG00000288658.1 5.777668e-01 6.693490e-01
ENSG00000288663.1 4.278680e-10 5.115145e-09
ENSG00000288670.1 8.125771e-02 1.371595e-01
ENSG00000288674.1 6.066099e-01 6.935602e-01
ENSG00000288675.1 8.599064e-22 4.931720e-20
[29995 rows x 6 columns]
Performing contrastive analysis for Her2 vs. Normal
Running Wald tests...
... done in 2.29 seconds.
Log2 fold change & Wald test p-value: paper_BRCA_Subtype_PAM50 Her2 vs Normal
baseMean log2FoldChange lfcSE stat \
ENSG00000000003.15 3087.392130 -0.103548 0.178483 -0.580154
ENSG00000000005.6 72.999641 -2.434328 0.473977 -5.135963
ENSG00000000419.13 2386.496112 0.656701 0.101685 6.458198
ENSG00000000457.14 1596.783158 0.522913 0.102761 5.088644
ENSG00000000460.17 715.140714 0.877735 0.131394 6.680153
... ... ... ... ...
ENSG00000288658.1 22.532026 0.220343 0.390278 0.564578
ENSG00000288663.1 29.867524 -0.521474 0.159972 -3.259780
ENSG00000288670.1 422.158728 0.551230 0.129099 4.269815
ENSG00000288674.1 8.197550 -0.443262 0.191417 -2.315687
ENSG00000288675.1 32.370671 0.572297 0.190145 3.009796
pvalue padj
ENSG00000000003.15 5.618107e-01 6.612585e-01
ENSG00000000005.6 2.807025e-07 2.722169e-06
ENSG00000000419.13 1.059569e-10 2.622258e-09
ENSG00000000457.14 3.606320e-07 3.381419e-06
ENSG00000000460.17 2.386925e-11 7.074684e-10
... ... ...
ENSG00000288658.1 5.723605e-01 6.706232e-01
ENSG00000288663.1 1.114985e-03 3.756484e-03
ENSG00000288670.1 1.956349e-05 1.138767e-04
ENSG00000288674.1 2.057536e-02 4.536925e-02
ENSG00000288675.1 2.614234e-03 7.828087e-03
[29995 rows x 6 columns]
Performing contrastive analysis for Her2 vs. Basal
Running Wald tests...
... done in 2.21 seconds.
Log2 fold change & Wald test p-value: paper_BRCA_Subtype_PAM50 Her2 vs Basal
baseMean log2FoldChange lfcSE stat \
ENSG00000000003.15 3087.392130 -0.399387 0.124227 -3.214986
ENSG00000000005.6 72.999641 -2.242245 0.331022 -6.773697
ENSG00000000419.13 2386.496112 0.101422 0.070733 1.433868
ENSG00000000457.14 1596.783158 0.512741 0.071486 7.172564
ENSG00000000460.17 715.140714 -0.581052 0.091224 -6.369509
... ... ... ... ...
ENSG00000288658.1 22.532026 -0.992407 0.270668 -3.666515
ENSG00000288663.1 29.867524 -0.038489 0.112749 -0.341369
ENSG00000288670.1 422.158728 0.117936 0.089665 1.315300
ENSG00000288674.1 8.197550 -0.255971 0.135260 -1.892439
ENSG00000288675.1 32.370671 0.584431 0.131657 4.439052
pvalue padj
ENSG00000000003.15 1.304511e-03 2.998375e-03
ENSG00000000005.6 1.255320e-11 8.772910e-11
ENSG00000000419.13 1.516099e-01 2.123529e-01
ENSG00000000457.14 7.360624e-13 5.828456e-12
ENSG00000000460.17 1.896341e-10 1.165828e-09
... ... ...
ENSG00000288658.1 2.458782e-04 6.493324e-04
ENSG00000288663.1 7.328255e-01 7.904312e-01
ENSG00000288670.1 1.884089e-01 2.559486e-01
ENSG00000288674.1 5.843248e-02 9.229501e-02
ENSG00000288675.1 9.035578e-06 3.005347e-05
[29995 rows x 6 columns]
Performing contrastive analysis for LumB vs. Normal
Running Wald tests...
... done in 2.46 seconds.
Log2 fold change & Wald test p-value: paper_BRCA_Subtype_PAM50 LumB vs Normal
baseMean log2FoldChange lfcSE stat \
ENSG00000000003.15 3087.392130 -0.660339 0.159710 -4.134599
ENSG00000000005.6 72.999641 -0.751062 0.422829 -1.776279
ENSG00000000419.13 2386.496112 0.548828 0.091000 6.031049
ENSG00000000457.14 1596.783158 0.389797 0.091967 4.238455
ENSG00000000460.17 715.140714 1.049255 0.117618 8.920861
... ... ... ... ...
ENSG00000288658.1 22.532026 0.072986 0.349326 0.208934
ENSG00000288663.1 29.867524 0.159890 0.141706 1.128321
ENSG00000288670.1 422.158728 0.703788 0.115554 6.090571
ENSG00000288674.1 8.197550 -0.375132 0.169207 -2.216998
ENSG00000288675.1 32.370671 -0.664767 0.171077 -3.885788
pvalue padj
ENSG00000000003.15 3.555757e-05 1.318795e-04
ENSG00000000005.6 7.568690e-02 1.186675e-01
ENSG00000000419.13 1.628993e-09 1.457548e-08
ENSG00000000457.14 2.250635e-05 8.702824e-05
ENSG00000000460.17 4.626757e-19 2.505046e-17
... ... ...
ENSG00000288658.1 8.345000e-01 8.730974e-01
ENSG00000288663.1 2.591845e-01 3.374382e-01
ENSG00000288670.1 1.125090e-09 1.042542e-08
ENSG00000288674.1 2.662322e-02 4.801079e-02
ENSG00000288675.1 1.019985e-04 3.432181e-04
[29995 rows x 6 columns]
Performing contrastive analysis for LumB vs. Basal
Running Wald tests...
... done in 2.19 seconds.
Log2 fold change & Wald test p-value: paper_BRCA_Subtype_PAM50 LumB vs Basal
baseMean log2FoldChange lfcSE stat \
ENSG00000000003.15 3087.392130 -0.956178 0.095308 -10.032555
ENSG00000000005.6 72.999641 -0.558979 0.252400 -2.214654
ENSG00000000419.13 2386.496112 -0.006451 0.054263 -0.118890
ENSG00000000457.14 1596.783158 0.379624 0.054849 6.921272
ENSG00000000460.17 715.140714 -0.409531 0.069938 -5.855629
... ... ... ... ...
ENSG00000288658.1 22.532026 -1.139764 0.207297 -5.498215
ENSG00000288663.1 29.867524 0.642875 0.084864 7.575379
ENSG00000288670.1 422.158728 0.270494 0.068745 3.934774
ENSG00000288674.1 8.197550 -0.187841 0.101419 -1.852129
ENSG00000288675.1 32.370671 -0.652634 0.102204 -6.385618
pvalue padj
ENSG00000000003.15 1.096424e-23 6.605189e-23
ENSG00000000005.6 2.678385e-02 3.592073e-02
ENSG00000000419.13 9.053625e-01 9.182818e-01
ENSG00000000457.14 4.476050e-12 1.394755e-11
ENSG00000000460.17 4.752078e-09 1.200628e-08
... ... ...
ENSG00000288658.1 3.836546e-08 9.064050e-08
ENSG00000288663.1 3.580819e-14 1.268982e-13
ENSG00000288670.1 8.327492e-05 1.475736e-04
ENSG00000288674.1 6.400724e-02 8.153584e-02
ENSG00000288675.1 1.707070e-10 4.801086e-10
[29995 rows x 6 columns]
Performing contrastive analysis for Normal vs. Basal
Running Wald tests...
... done in 2.34 seconds.
Log2 fold change & Wald test p-value: paper_BRCA_Subtype_PAM50 Normal vs Basal
baseMean log2FoldChange lfcSE stat \
ENSG00000000003.15 3087.392130 -0.295840 0.161945 -1.826789
ENSG00000000005.6 72.999641 0.192083 0.428710 0.448048
ENSG00000000419.13 2386.496112 -0.555279 0.092278 -6.017474
ENSG00000000457.14 1596.783158 -0.010172 0.093272 -0.109060
ENSG00000000460.17 715.140714 -1.458786 0.119249 -12.233141
... ... ... ... ...
ENSG00000288658.1 22.532026 -1.212750 0.353853 -3.427268
ENSG00000288663.1 29.867524 0.482985 0.144510 3.342218
ENSG00000288670.1 422.158728 -0.433294 0.117198 -3.697105
ENSG00000288674.1 8.197550 0.187291 0.171522 1.091936
ENSG00000288675.1 32.370671 0.012134 0.173117 0.070089
pvalue padj
ENSG00000000003.15 6.773147e-02 1.155634e-01
ENSG00000000005.6 6.541185e-01 7.328111e-01
ENSG00000000419.13 1.771600e-09 1.772487e-08
ENSG00000000457.14 9.131548e-01 9.377274e-01
ENSG00000000460.17 2.067839e-34 5.743039e-32
... ... ...
ENSG00000288658.1 6.096865e-04 1.915328e-03
ENSG00000288663.1 8.311184e-04 2.519648e-03
ENSG00000288670.1 2.180718e-04 7.638751e-04
ENSG00000288674.1 2.748612e-01 3.696571e-01
ENSG00000288675.1 9.441229e-01 9.600946e-01
[29995 rows x 6 columns]
Fit type used for VST : parametric
Fitting dispersions...
... done in 9.30 seconds.
Clearing gpu memory
Total = 42.4Gb Reserved = 0.0Gb Allocated = 0.0Gb
GCN_MME(
(encoder_dims): ModuleList(
(0): Encoder(
(encoder): ModuleList(
(0): Linear(in_features=464, out_features=500, bias=True)
(1): Linear(in_features=500, out_features=16, bias=True)
)
(norm): ModuleList(
(0): BatchNorm1d(500, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(1): BatchNorm1d(16, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
)
(decoder): Sequential(
(0): Linear(in_features=16, out_features=64, bias=True)
)
(drop): Dropout(p=0.5, inplace=False)
)
(1): Encoder(
(encoder): ModuleList(
(0): Linear(in_features=29995, out_features=500, bias=True)
(1): Linear(in_features=500, out_features=16, bias=True)
)
(norm): ModuleList(
(0): BatchNorm1d(500, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(1): BatchNorm1d(16, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
)
(decoder): Sequential(
(0): Linear(in_features=16, out_features=64, bias=True)
)
(drop): Dropout(p=0.5, inplace=False)
)
)
(gnnlayers): ModuleList(
(0): GraphConv(in=64, out=32, normalization=both, activation=None)
(1): GraphConv(in=32, out=5, normalization=both, activation=None)
)
(batch_norms): ModuleList(
(0): BatchNorm1d(32, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
)
(drop): Dropout(p=0.5, inplace=False)
)
Graph(num_nodes=1076, num_edges=18340,
ndata_schemes={'idx': Scheme(shape=(), dtype=torch.int64), 'label': Scheme(shape=(5,), dtype=torch.int64), 'feat': Scheme(shape=(30459,), dtype=torch.float32)}
edata_schemes={})
Epoch 00000 | Loss 1.8291 | Train Acc. 0.2230 |
Epoch 00005 | Loss 0.6409 | Train Acc. 0.7974 |
Epoch 00010 | Loss 0.5584 | Train Acc. 0.8309 |
Epoch 00015 | Loss 0.4914 | Train Acc. 0.8383 |
Epoch 00020 | Loss 0.4561 | Train Acc. 0.8457 |
Epoch 00025 | Loss 0.4449 | Train Acc. 0.8327 |
Epoch 00030 | Loss 0.4324 | Train Acc. 0.8309 |
Epoch 00035 | Loss 0.3926 | Train Acc. 0.8494 |
Epoch 00040 | Loss 0.4163 | Train Acc. 0.8569 |
Epoch 00045 | Loss 0.3781 | Train Acc. 0.8532 |
Epoch 00050 | Loss 0.3679 | Train Acc. 0.8550 |
Epoch 00055 | Loss 0.3676 | Train Acc. 0.8439 |
Epoch 00060 | Loss 0.3519 | Train Acc. 0.8662 |
Epoch 00065 | Loss 0.3529 | Train Acc. 0.8643 |
Epoch 00070 | Loss 0.3619 | Train Acc. 0.8643 |
Epoch 00075 | Loss 0.3438 | Train Acc. 0.8662 |
Epoch 00080 | Loss 0.3323 | Train Acc. 0.8717 |
Epoch 00085 | Loss 0.3296 | Train Acc. 0.8755 |
Epoch 00090 | Loss 0.3310 | Train Acc. 0.8736 |
Epoch 00095 | Loss 0.3275 | Train Acc. 0.8736 |
Epoch 00100 | Loss 0.3142 | Train Acc. 0.8717 |
Epoch 00105 | Loss 0.3110 | Train Acc. 0.8792 |
Epoch 00110 | Loss 0.3265 | Train Acc. 0.8662 |
Epoch 00115 | Loss 0.3329 | Train Acc. 0.8625 |
Epoch 00120 | Loss 0.3043 | Train Acc. 0.8866 |
Epoch 00125 | Loss 0.3079 | Train Acc. 0.8792 |
Epoch 00130 | Loss 0.2987 | Train Acc. 0.8810 |
Epoch 00135 | Loss 0.3162 | Train Acc. 0.8773 |
Epoch 00140 | Loss 0.3199 | Train Acc. 0.8755 |
Epoch 00145 | Loss 0.3026 | Train Acc. 0.8792 |
Epoch 00150 | Loss 0.3020 | Train Acc. 0.8755 |
Epoch 00155 | Loss 0.3111 | Train Acc. 0.8773 |
Epoch 00160 | Loss 0.3019 | Train Acc. 0.8866 |
Epoch 00165 | Loss 0.3063 | Train Acc. 0.8903 |
Epoch 00170 | Loss 0.2964 | Train Acc. 0.8773 |
Epoch 00175 | Loss 0.2888 | Train Acc. 0.8885 |
Epoch 00180 | Loss 0.2945 | Train Acc. 0.8922 |
Epoch 00185 | Loss 0.2923 | Train Acc. 0.8996 |
Epoch 00190 | Loss 0.2908 | Train Acc. 0.8922 |
Epoch 00195 | Loss 0.2977 | Train Acc. 0.8903 |
Epoch 00200 | Loss 0.2945 | Train Acc. 0.8848 |
Epoch 00205 | Loss 0.2964 | Train Acc. 0.8810 |
Epoch 00210 | Loss 0.2782 | Train Acc. 0.8996 |
Epoch 00215 | Loss 0.2982 | Train Acc. 0.8680 |
Epoch 00220 | Loss 0.2880 | Train Acc. 0.8848 |
Epoch 00225 | Loss 0.2932 | Train Acc. 0.8866 |
Epoch 00230 | Loss 0.2993 | Train Acc. 0.8773 |
Epoch 00235 | Loss 0.2817 | Train Acc. 0.8978 |
Epoch 00240 | Loss 0.2857 | Train Acc. 0.8866 |
Epoch 00245 | Loss 0.2928 | Train Acc. 0.8941 |
Epoch 00250 | Loss 0.2903 | Train Acc. 0.8810 |
Epoch 00255 | Loss 0.2977 | Train Acc. 0.8866 |
Epoch 00260 | Loss 0.3070 | Train Acc. 0.8792 |
Epoch 00265 | Loss 0.3077 | Train Acc. 0.8922 |
Epoch 00270 | Loss 0.2879 | Train Acc. 0.8885 |
Epoch 00275 | Loss 0.2780 | Train Acc. 0.8829 |
Epoch 00280 | Loss 0.2829 | Train Acc. 0.8829 |
Epoch 00285 | Loss 0.2986 | Train Acc. 0.8755 |
Epoch 00290 | Loss 0.2913 | Train Acc. 0.8922 |
Epoch 00295 | Loss 0.2870 | Train Acc. 0.8996 |
Epoch 00300 | Loss 0.2759 | Train Acc. 0.8866 |
Epoch 00305 | Loss 0.2921 | Train Acc. 0.8903 |
Epoch 00310 | Loss 0.2996 | Train Acc. 0.8941 |
Epoch 00315 | Loss 0.2841 | Train Acc. 0.8885 |
Epoch 00320 | Loss 0.3018 | Train Acc. 0.8903 |
Epoch 00325 | Loss 0.2918 | Train Acc. 0.8903 |
Epoch 00330 | Loss 0.2901 | Train Acc. 0.8829 |
Epoch 00335 | Loss 0.2753 | Train Acc. 0.8866 |
Epoch 00340 | Loss 0.2861 | Train Acc. 0.8885 |
Epoch 00345 | Loss 0.2779 | Train Acc. 0.8922 |
Epoch 00350 | Loss 0.2898 | Train Acc. 0.8922 |
Epoch 00355 | Loss 0.2846 | Train Acc. 0.9015 |
Epoch 00360 | Loss 0.2750 | Train Acc. 0.8959 |
Epoch 00365 | Loss 0.2774 | Train Acc. 0.8885 |
Epoch 00370 | Loss 0.2826 | Train Acc. 0.8885 |
Epoch 00375 | Loss 0.2694 | Train Acc. 0.8922 |
Epoch 00380 | Loss 0.3013 | Train Acc. 0.8773 |
Epoch 00385 | Loss 0.2818 | Train Acc. 0.8903 |
Epoch 00390 | Loss 0.2829 | Train Acc. 0.8866 |
Epoch 00395 | Loss 0.2822 | Train Acc. 0.8885 |
Epoch 00400 | Loss 0.2829 | Train Acc. 0.8903 |
Epoch 00405 | Loss 0.2885 | Train Acc. 0.8848 |
Epoch 00410 | Loss 0.2770 | Train Acc. 0.8922 |
Epoch 00415 | Loss 0.2764 | Train Acc. 0.8959 |
Epoch 00420 | Loss 0.2841 | Train Acc. 0.8885 |
Epoch 00425 | Loss 0.2682 | Train Acc. 0.8978 |
Epoch 00430 | Loss 0.2932 | Train Acc. 0.8773 |
Epoch 00435 | Loss 0.2790 | Train Acc. 0.8922 |
Epoch 00440 | Loss 0.2649 | Train Acc. 0.8848 |
Epoch 00445 | Loss 0.3039 | Train Acc. 0.8866 |
Epoch 00450 | Loss 0.2829 | Train Acc. 0.8829 |
Epoch 00455 | Loss 0.2901 | Train Acc. 0.8829 |
Epoch 00460 | Loss 0.2908 | Train Acc. 0.8792 |
Epoch 00465 | Loss 0.2677 | Train Acc. 0.8941 |
Epoch 00470 | Loss 0.2917 | Train Acc. 0.8978 |
Early stopping! No improvement for 100 consecutive epochs.
Fold : 1 | Test Accuracy = 0.8364 | F1 = 0.8039
Total = 42.4Gb Reserved = 0.6Gb Allocated = 0.3Gb
Clearing gpu memory
Total = 42.4Gb Reserved = 0.3Gb Allocated = 0.3Gb
Performing Logistic Regression for Feature Selection
Loss : 0.6041: 100%|██████████| 1000/1000 [00:02<00:00, 406.47epoch/s]
Model score : 0.877
<Figure size 640x480 with 0 Axes>
Performing Differential Gene Expression for Feature Selection
Keeping 29995 genes
Removed 30665 genes
Keeping 1047 Samples
Removed 36 Samples
Fitting size factors...
Using None as control genes, passed at DeseqDataSet initialization
... done in 1.19 seconds.
Fitting dispersions...
... done in 10.30 seconds.
Fitting dispersion trend curve...
... done in 0.59 seconds.
Fitting MAP dispersions...
... done in 11.29 seconds.
Fitting LFCs...
... done in 6.66 seconds.
Calculating cook's distance...
... done in 2.13 seconds.
Replacing 2948 outlier genes.
Fitting dispersions...
... done in 1.10 seconds.
Fitting MAP dispersions...
... done in 1.05 seconds.
Fitting LFCs...
... done in 1.00 seconds.
Performing contrastive analysis for LumA vs. Her2
Running Wald tests...
... done in 2.68 seconds.
Log2 fold change & Wald test p-value: paper_BRCA_Subtype_PAM50 LumA vs Her2
baseMean log2FoldChange lfcSE stat \
ENSG00000000003.15 3087.392130 -0.045843 0.109944 -0.416972
ENSG00000000005.6 72.999641 2.364056 0.293465 8.055675
ENSG00000000419.13 2386.496112 -0.604066 0.062602 -9.649254
ENSG00000000457.14 1596.783158 -0.062327 0.063247 -0.985452
ENSG00000000460.17 715.140714 -0.435593 0.080773 -5.392793
... ... ... ... ...
ENSG00000288658.1 22.532026 -0.955682 0.240269 -3.977559
ENSG00000288663.1 29.867524 0.627702 0.099567 6.304313
ENSG00000288670.1 422.158728 -0.075705 0.079346 -0.954105
ENSG00000288674.1 8.197550 0.337858 0.120301 2.808436
ENSG00000288675.1 32.370671 -0.912699 0.116411 -7.840287
pvalue padj
ENSG00000000003.15 6.766989e-01 7.320512e-01
ENSG00000000005.6 7.904166e-16 9.221527e-15
ENSG00000000419.13 4.951465e-22 1.092053e-20
ENSG00000000457.14 3.244019e-01 3.944238e-01
ENSG00000000460.17 6.937103e-08 3.022199e-07
... ... ...
ENSG00000288658.1 6.962648e-05 1.878268e-04
ENSG00000288663.1 2.894748e-10 1.756229e-09
ENSG00000288670.1 3.400305e-01 4.104478e-01
ENSG00000288674.1 4.978277e-03 9.501362e-03
ENSG00000288675.1 4.495165e-15 4.869356e-14
[29995 rows x 6 columns]
Performing contrastive analysis for LumA vs. LumB
Running Wald tests...
... done in 2.66 seconds.
Log2 fold change & Wald test p-value: paper_BRCA_Subtype_PAM50 LumA vs LumB
baseMean log2FoldChange lfcSE stat \
ENSG00000000003.15 3087.392130 0.510947 0.075755 6.744707
ENSG00000000005.6 72.999641 0.680790 0.200628 3.393294
ENSG00000000419.13 2386.496112 -0.496193 0.043131 -11.504238
ENSG00000000457.14 1596.783158 0.070789 0.043570 1.624718
ENSG00000000460.17 715.140714 -0.607113 0.055622 -10.914917
... ... ... ... ...
ENSG00000288658.1 22.532026 -0.808325 0.165651 -4.879702
ENSG00000288663.1 29.867524 -0.053662 0.066356 -0.808701
ENSG00000288670.1 422.158728 -0.228263 0.054607 -4.180137
ENSG00000288674.1 8.197550 0.269729 0.080393 3.355135
ENSG00000288675.1 32.370671 0.324366 0.081632 3.973538
pvalue padj
ENSG00000000003.15 1.533361e-11 1.663405e-10
ENSG00000000005.6 6.905757e-04 1.840247e-03
ENSG00000000419.13 1.255933e-30 1.034937e-28
ENSG00000000457.14 1.042227e-01 1.570698e-01
ENSG00000000460.17 9.781815e-28 6.378381e-26
... ... ...
ENSG00000288658.1 1.062460e-06 5.130976e-06
ENSG00000288663.1 4.186870e-01 5.071215e-01
ENSG00000288670.1 2.913337e-05 1.041544e-04
ENSG00000288674.1 7.932621e-04 2.084624e-03
ENSG00000288675.1 7.081273e-05 2.341559e-04
[29995 rows x 6 columns]
Performing contrastive analysis for LumA vs. Normal
Running Wald tests...
... done in 2.08 seconds.
Log2 fold change & Wald test p-value: paper_BRCA_Subtype_PAM50 LumA vs Normal
baseMean log2FoldChange lfcSE stat \
ENSG00000000003.15 3087.392130 -0.149391 0.151267 -0.987602
ENSG00000000005.6 72.999641 -0.070272 0.400423 -0.175495
ENSG00000000419.13 2386.496112 0.052635 0.086204 0.610591
ENSG00000000457.14 1596.783158 0.460586 0.087118 5.286911
ENSG00000000460.17 715.140714 0.442141 0.111457 3.966908
... ... ... ... ...
ENSG00000288658.1 22.532026 -0.735339 0.331180 -2.220363
ENSG00000288663.1 29.867524 0.106228 0.134479 0.789924
ENSG00000288670.1 422.158728 0.475525 0.109506 4.342477
ENSG00000288674.1 8.197550 -0.105403 0.159991 -0.658810
ENSG00000288675.1 32.370671 -0.340401 0.161826 -2.103501
pvalue padj
ENSG00000000003.15 3.233477e-01 0.443638
ENSG00000000005.6 8.606909e-01 0.906762
ENSG00000000419.13 5.414706e-01 0.651681
ENSG00000000457.14 1.243991e-07 0.000002
ENSG00000000460.17 7.281114e-05 0.000401
... ... ...
ENSG00000288658.1 2.639415e-02 0.059055
ENSG00000288663.1 4.295720e-01 0.548929
ENSG00000288670.1 1.408855e-05 0.000097
ENSG00000288674.1 5.100176e-01 0.624223
ENSG00000288675.1 3.542197e-02 0.075375
[29995 rows x 6 columns]
Performing contrastive analysis for LumA vs. Basal
Running Wald tests...
... done in 2.11 seconds.
Log2 fold change & Wald test p-value: paper_BRCA_Subtype_PAM50 LumA vs Basal
baseMean log2FoldChange lfcSE stat \
ENSG00000000003.15 3087.392130 -0.445231 0.080360 -5.540469
ENSG00000000005.6 72.999641 0.121811 0.212744 0.572570
ENSG00000000419.13 2386.496112 -0.502644 0.045765 -10.983180
ENSG00000000457.14 1596.783158 0.450413 0.046261 9.736320
ENSG00000000460.17 715.140714 -1.016645 0.058992 -17.233544
... ... ... ... ...
ENSG00000288658.1 22.532026 -1.948089 0.174996 -11.132162
ENSG00000288663.1 29.867524 0.589213 0.072151 8.166402
ENSG00000288670.1 422.158728 0.042231 0.058005 0.728061
ENSG00000288674.1 8.197550 0.081888 0.085157 0.961606
ENSG00000288675.1 32.370671 -0.328268 0.085826 -3.824821
pvalue padj
ENSG00000000003.15 3.016618e-08 5.818872e-08
ENSG00000000005.6 5.669360e-01 6.009346e-01
ENSG00000000419.13 4.604242e-28 2.082392e-27
ENSG00000000457.14 2.110588e-22 7.768694e-22
ENSG00000000460.17 1.487504e-66 1.983009e-65
... ... ...
ENSG00000288658.1 8.748769e-29 4.064106e-28
ENSG00000288663.1 3.177226e-16 9.097937e-16
ENSG00000288670.1 4.665759e-01 5.029268e-01
ENSG00000288674.1 3.362475e-01 3.704861e-01
ENSG00000288675.1 1.308669e-04 2.016518e-04
[29995 rows x 6 columns]
Performing contrastive analysis for Her2 vs. LumB
Running Wald tests...
... done in 2.42 seconds.
Log2 fold change & Wald test p-value: paper_BRCA_Subtype_PAM50 Her2 vs LumB
baseMean log2FoldChange lfcSE stat \
ENSG00000000003.15 3087.392130 0.556791 0.121299 4.590230
ENSG00000000005.6 72.999641 -1.683266 0.323369 -5.205406
ENSG00000000419.13 2386.496112 0.107873 0.069059 1.562059
ENSG00000000457.14 1596.783158 0.133116 0.069775 1.907792
ENSG00000000460.17 715.140714 -0.171520 0.089082 -1.925423
... ... ... ... ...
ENSG00000288658.1 22.532026 0.147357 0.264721 0.556650
ENSG00000288663.1 29.867524 -0.681364 0.109132 -6.243507
ENSG00000288670.1 422.158728 -0.152558 0.087504 -1.743435
ENSG00000288674.1 8.197550 -0.068130 0.132312 -0.514919
ENSG00000288675.1 32.370671 1.237065 0.128962 9.592483
pvalue padj
ENSG00000000003.15 4.427577e-06 2.483732e-05
ENSG00000000005.6 1.935725e-07 1.438248e-06
ENSG00000000419.13 1.182741e-01 1.871212e-01
ENSG00000000457.14 5.641806e-02 1.010847e-01
ENSG00000000460.17 5.417652e-02 9.782234e-02
... ... ...
ENSG00000288658.1 5.777668e-01 6.693490e-01
ENSG00000288663.1 4.278680e-10 5.115145e-09
ENSG00000288670.1 8.125771e-02 1.371595e-01
ENSG00000288674.1 6.066099e-01 6.935602e-01
ENSG00000288675.1 8.599064e-22 4.931720e-20
[29995 rows x 6 columns]
Performing contrastive analysis for Her2 vs. Normal
Running Wald tests...
... done in 2.60 seconds.
Log2 fold change & Wald test p-value: paper_BRCA_Subtype_PAM50 Her2 vs Normal
baseMean log2FoldChange lfcSE stat \
ENSG00000000003.15 3087.392130 -0.103548 0.178483 -0.580154
ENSG00000000005.6 72.999641 -2.434328 0.473977 -5.135963
ENSG00000000419.13 2386.496112 0.656701 0.101685 6.458198
ENSG00000000457.14 1596.783158 0.522913 0.102761 5.088644
ENSG00000000460.17 715.140714 0.877735 0.131394 6.680153
... ... ... ... ...
ENSG00000288658.1 22.532026 0.220343 0.390278 0.564578
ENSG00000288663.1 29.867524 -0.521474 0.159972 -3.259780
ENSG00000288670.1 422.158728 0.551230 0.129099 4.269815
ENSG00000288674.1 8.197550 -0.443262 0.191417 -2.315687
ENSG00000288675.1 32.370671 0.572297 0.190145 3.009796
pvalue padj
ENSG00000000003.15 5.618107e-01 6.612585e-01
ENSG00000000005.6 2.807025e-07 2.722169e-06
ENSG00000000419.13 1.059569e-10 2.622258e-09
ENSG00000000457.14 3.606320e-07 3.381419e-06
ENSG00000000460.17 2.386925e-11 7.074684e-10
... ... ...
ENSG00000288658.1 5.723605e-01 6.706232e-01
ENSG00000288663.1 1.114985e-03 3.756484e-03
ENSG00000288670.1 1.956349e-05 1.138767e-04
ENSG00000288674.1 2.057536e-02 4.536925e-02
ENSG00000288675.1 2.614234e-03 7.828087e-03
[29995 rows x 6 columns]
Performing contrastive analysis for Her2 vs. Basal
Running Wald tests...
... done in 2.37 seconds.
Log2 fold change & Wald test p-value: paper_BRCA_Subtype_PAM50 Her2 vs Basal
baseMean log2FoldChange lfcSE stat \
ENSG00000000003.15 3087.392130 -0.399387 0.124227 -3.214986
ENSG00000000005.6 72.999641 -2.242245 0.331022 -6.773697
ENSG00000000419.13 2386.496112 0.101422 0.070733 1.433868
ENSG00000000457.14 1596.783158 0.512741 0.071486 7.172564
ENSG00000000460.17 715.140714 -0.581052 0.091224 -6.369509
... ... ... ... ...
ENSG00000288658.1 22.532026 -0.992407 0.270668 -3.666515
ENSG00000288663.1 29.867524 -0.038489 0.112749 -0.341369
ENSG00000288670.1 422.158728 0.117936 0.089665 1.315300
ENSG00000288674.1 8.197550 -0.255971 0.135260 -1.892439
ENSG00000288675.1 32.370671 0.584431 0.131657 4.439052
pvalue padj
ENSG00000000003.15 1.304511e-03 2.998375e-03
ENSG00000000005.6 1.255320e-11 8.772910e-11
ENSG00000000419.13 1.516099e-01 2.123529e-01
ENSG00000000457.14 7.360624e-13 5.828456e-12
ENSG00000000460.17 1.896341e-10 1.165828e-09
... ... ...
ENSG00000288658.1 2.458782e-04 6.493324e-04
ENSG00000288663.1 7.328255e-01 7.904312e-01
ENSG00000288670.1 1.884089e-01 2.559486e-01
ENSG00000288674.1 5.843248e-02 9.229501e-02
ENSG00000288675.1 9.035578e-06 3.005347e-05
[29995 rows x 6 columns]
Performing contrastive analysis for LumB vs. Normal
Running Wald tests...
... done in 2.62 seconds.
Log2 fold change & Wald test p-value: paper_BRCA_Subtype_PAM50 LumB vs Normal
baseMean log2FoldChange lfcSE stat \
ENSG00000000003.15 3087.392130 -0.660339 0.159710 -4.134599
ENSG00000000005.6 72.999641 -0.751062 0.422829 -1.776279
ENSG00000000419.13 2386.496112 0.548828 0.091000 6.031049
ENSG00000000457.14 1596.783158 0.389797 0.091967 4.238455
ENSG00000000460.17 715.140714 1.049255 0.117618 8.920861
... ... ... ... ...
ENSG00000288658.1 22.532026 0.072986 0.349326 0.208934
ENSG00000288663.1 29.867524 0.159890 0.141706 1.128321
ENSG00000288670.1 422.158728 0.703788 0.115554 6.090571
ENSG00000288674.1 8.197550 -0.375132 0.169207 -2.216998
ENSG00000288675.1 32.370671 -0.664767 0.171077 -3.885788
pvalue padj
ENSG00000000003.15 3.555757e-05 1.318795e-04
ENSG00000000005.6 7.568690e-02 1.186675e-01
ENSG00000000419.13 1.628993e-09 1.457548e-08
ENSG00000000457.14 2.250635e-05 8.702824e-05
ENSG00000000460.17 4.626757e-19 2.505046e-17
... ... ...
ENSG00000288658.1 8.345000e-01 8.730974e-01
ENSG00000288663.1 2.591845e-01 3.374382e-01
ENSG00000288670.1 1.125090e-09 1.042542e-08
ENSG00000288674.1 2.662322e-02 4.801079e-02
ENSG00000288675.1 1.019985e-04 3.432181e-04
[29995 rows x 6 columns]
Performing contrastive analysis for LumB vs. Basal
Running Wald tests...
... done in 2.88 seconds.
Log2 fold change & Wald test p-value: paper_BRCA_Subtype_PAM50 LumB vs Basal
baseMean log2FoldChange lfcSE stat \
ENSG00000000003.15 3087.392130 -0.956178 0.095308 -10.032555
ENSG00000000005.6 72.999641 -0.558979 0.252400 -2.214654
ENSG00000000419.13 2386.496112 -0.006451 0.054263 -0.118890
ENSG00000000457.14 1596.783158 0.379624 0.054849 6.921272
ENSG00000000460.17 715.140714 -0.409531 0.069938 -5.855629
... ... ... ... ...
ENSG00000288658.1 22.532026 -1.139764 0.207297 -5.498215
ENSG00000288663.1 29.867524 0.642875 0.084864 7.575379
ENSG00000288670.1 422.158728 0.270494 0.068745 3.934774
ENSG00000288674.1 8.197550 -0.187841 0.101419 -1.852129
ENSG00000288675.1 32.370671 -0.652634 0.102204 -6.385618
pvalue padj
ENSG00000000003.15 1.096424e-23 6.605189e-23
ENSG00000000005.6 2.678385e-02 3.592073e-02
ENSG00000000419.13 9.053625e-01 9.182818e-01
ENSG00000000457.14 4.476050e-12 1.394755e-11
ENSG00000000460.17 4.752078e-09 1.200628e-08
... ... ...
ENSG00000288658.1 3.836546e-08 9.064050e-08
ENSG00000288663.1 3.580819e-14 1.268982e-13
ENSG00000288670.1 8.327492e-05 1.475736e-04
ENSG00000288674.1 6.400724e-02 8.153584e-02
ENSG00000288675.1 1.707070e-10 4.801086e-10
[29995 rows x 6 columns]
Performing contrastive analysis for Normal vs. Basal
Running Wald tests...
... done in 2.37 seconds.
Log2 fold change & Wald test p-value: paper_BRCA_Subtype_PAM50 Normal vs Basal
baseMean log2FoldChange lfcSE stat \
ENSG00000000003.15 3087.392130 -0.295840 0.161945 -1.826789
ENSG00000000005.6 72.999641 0.192083 0.428710 0.448048
ENSG00000000419.13 2386.496112 -0.555279 0.092278 -6.017474
ENSG00000000457.14 1596.783158 -0.010172 0.093272 -0.109060
ENSG00000000460.17 715.140714 -1.458786 0.119249 -12.233141
... ... ... ... ...
ENSG00000288658.1 22.532026 -1.212750 0.353853 -3.427268
ENSG00000288663.1 29.867524 0.482985 0.144510 3.342218
ENSG00000288670.1 422.158728 -0.433294 0.117198 -3.697105
ENSG00000288674.1 8.197550 0.187291 0.171522 1.091936
ENSG00000288675.1 32.370671 0.012134 0.173117 0.070089
pvalue padj
ENSG00000000003.15 6.773147e-02 1.155634e-01
ENSG00000000005.6 6.541185e-01 7.328111e-01
ENSG00000000419.13 1.771600e-09 1.772487e-08
ENSG00000000457.14 9.131548e-01 9.377274e-01
ENSG00000000460.17 2.067839e-34 5.743039e-32
... ... ...
ENSG00000288658.1 6.096865e-04 1.915328e-03
ENSG00000288663.1 8.311184e-04 2.519648e-03
ENSG00000288670.1 2.180718e-04 7.638751e-04
ENSG00000288674.1 2.748612e-01 3.696571e-01
ENSG00000288675.1 9.441229e-01 9.600946e-01
[29995 rows x 6 columns]
Fit type used for VST : parametric
Fitting dispersions...
... done in 9.63 seconds.
Clearing gpu memory
Total = 42.4Gb Reserved = 0.3Gb Allocated = 0.3Gb

GCN_MME(
(encoder_dims): ModuleList(
(0): Encoder(
(encoder): ModuleList(
(0): Linear(in_features=464, out_features=500, bias=True)
(1): Linear(in_features=500, out_features=16, bias=True)
)
(norm): ModuleList(
(0): BatchNorm1d(500, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(1): BatchNorm1d(16, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
)
(decoder): Sequential(
(0): Linear(in_features=16, out_features=64, bias=True)
)
(drop): Dropout(p=0.5, inplace=False)
)
(1): Encoder(
(encoder): ModuleList(
(0): Linear(in_features=29995, out_features=500, bias=True)
(1): Linear(in_features=500, out_features=16, bias=True)
)
(norm): ModuleList(
(0): BatchNorm1d(500, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(1): BatchNorm1d(16, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
)
(decoder): Sequential(
(0): Linear(in_features=16, out_features=64, bias=True)
)
(drop): Dropout(p=0.5, inplace=False)
)
)
(gnnlayers): ModuleList(
(0): GraphConv(in=64, out=32, normalization=both, activation=None)
(1): GraphConv(in=32, out=5, normalization=both, activation=None)
)
(batch_norms): ModuleList(
(0): BatchNorm1d(32, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
)
(drop): Dropout(p=0.5, inplace=False)
)
Graph(num_nodes=1076, num_edges=18346,
ndata_schemes={'idx': Scheme(shape=(), dtype=torch.int64), 'label': Scheme(shape=(5,), dtype=torch.int64), 'feat': Scheme(shape=(30459,), dtype=torch.float32)}
edata_schemes={})
Epoch 00000 | Loss 1.8456 | Train Acc. 0.2435 |
Epoch 00005 | Loss 0.5908 | Train Acc. 0.8030 |
Epoch 00010 | Loss 0.5297 | Train Acc. 0.8197 |
Epoch 00015 | Loss 0.5006 | Train Acc. 0.8178 |
Epoch 00020 | Loss 0.4776 | Train Acc. 0.8327 |
Epoch 00025 | Loss 0.4348 | Train Acc. 0.8494 |
Epoch 00030 | Loss 0.4500 | Train Acc. 0.8309 |
Epoch 00035 | Loss 0.4246 | Train Acc. 0.8364 |
Epoch 00040 | Loss 0.4182 | Train Acc. 0.8439 |
Epoch 00045 | Loss 0.4020 | Train Acc. 0.8532 |
Epoch 00050 | Loss 0.3798 | Train Acc. 0.8439 |
Epoch 00055 | Loss 0.4173 | Train Acc. 0.8439 |
Epoch 00060 | Loss 0.3841 | Train Acc. 0.8625 |
Epoch 00065 | Loss 0.3747 | Train Acc. 0.8532 |
Epoch 00070 | Loss 0.3782 | Train Acc. 0.8550 |
Epoch 00075 | Loss 0.3646 | Train Acc. 0.8662 |
Epoch 00080 | Loss 0.3678 | Train Acc. 0.8513 |
Epoch 00085 | Loss 0.3664 | Train Acc. 0.8550 |
Epoch 00090 | Loss 0.3517 | Train Acc. 0.8717 |
Epoch 00095 | Loss 0.3592 | Train Acc. 0.8606 |
Epoch 00100 | Loss 0.3514 | Train Acc. 0.8606 |
Epoch 00105 | Loss 0.3428 | Train Acc. 0.8625 |
Epoch 00110 | Loss 0.3339 | Train Acc. 0.8680 |
Epoch 00115 | Loss 0.3399 | Train Acc. 0.8699 |
Epoch 00120 | Loss 0.3467 | Train Acc. 0.8699 |
Epoch 00125 | Loss 0.3423 | Train Acc. 0.8755 |
Epoch 00130 | Loss 0.3418 | Train Acc. 0.8680 |
Epoch 00135 | Loss 0.3335 | Train Acc. 0.8792 |
Epoch 00140 | Loss 0.3254 | Train Acc. 0.8755 |
Epoch 00145 | Loss 0.3189 | Train Acc. 0.8755 |
Epoch 00150 | Loss 0.3212 | Train Acc. 0.8736 |
Epoch 00155 | Loss 0.3202 | Train Acc. 0.8717 |
Epoch 00160 | Loss 0.3114 | Train Acc. 0.8736 |
Epoch 00165 | Loss 0.3391 | Train Acc. 0.8736 |
Epoch 00170 | Loss 0.3198 | Train Acc. 0.8755 |
Epoch 00175 | Loss 0.3160 | Train Acc. 0.8717 |
Epoch 00180 | Loss 0.3202 | Train Acc. 0.8736 |
Epoch 00185 | Loss 0.3093 | Train Acc. 0.8829 |
Epoch 00190 | Loss 0.3128 | Train Acc. 0.8736 |
Epoch 00195 | Loss 0.3171 | Train Acc. 0.8829 |
Epoch 00200 | Loss 0.2978 | Train Acc. 0.8792 |
Epoch 00205 | Loss 0.3125 | Train Acc. 0.8625 |
Epoch 00210 | Loss 0.3087 | Train Acc. 0.8885 |
Epoch 00215 | Loss 0.3027 | Train Acc. 0.8829 |
Epoch 00220 | Loss 0.2999 | Train Acc. 0.8829 |
Epoch 00225 | Loss 0.3013 | Train Acc. 0.8848 |
Epoch 00230 | Loss 0.2968 | Train Acc. 0.8903 |
Epoch 00235 | Loss 0.3221 | Train Acc. 0.8848 |
Epoch 00240 | Loss 0.3107 | Train Acc. 0.8773 |
Epoch 00245 | Loss 0.2929 | Train Acc. 0.8959 |
Epoch 00250 | Loss 0.2958 | Train Acc. 0.8792 |
Epoch 00255 | Loss 0.2954 | Train Acc. 0.8848 |
Epoch 00260 | Loss 0.2980 | Train Acc. 0.8922 |
Epoch 00265 | Loss 0.3129 | Train Acc. 0.8810 |
Epoch 00270 | Loss 0.3026 | Train Acc. 0.8829 |
Epoch 00275 | Loss 0.2947 | Train Acc. 0.8866 |
Epoch 00280 | Loss 0.3082 | Train Acc. 0.8829 |
Epoch 00285 | Loss 0.3050 | Train Acc. 0.8792 |
Epoch 00290 | Loss 0.3064 | Train Acc. 0.8680 |
Epoch 00295 | Loss 0.3073 | Train Acc. 0.8755 |
Epoch 00300 | Loss 0.3084 | Train Acc. 0.8792 |
Epoch 00305 | Loss 0.2885 | Train Acc. 0.8978 |
Epoch 00310 | Loss 0.2965 | Train Acc. 0.8829 |
Epoch 00315 | Loss 0.3088 | Train Acc. 0.8662 |
Epoch 00320 | Loss 0.3027 | Train Acc. 0.8903 |
Epoch 00325 | Loss 0.3060 | Train Acc. 0.8848 |
Epoch 00330 | Loss 0.3038 | Train Acc. 0.8885 |
Epoch 00335 | Loss 0.3032 | Train Acc. 0.8736 |
Epoch 00340 | Loss 0.2948 | Train Acc. 0.8810 |
Epoch 00345 | Loss 0.3166 | Train Acc. 0.8810 |
Epoch 00350 | Loss 0.3035 | Train Acc. 0.8885 |
Epoch 00355 | Loss 0.2869 | Train Acc. 0.8959 |
Epoch 00360 | Loss 0.3090 | Train Acc. 0.8810 |
Epoch 00365 | Loss 0.3108 | Train Acc. 0.8848 |
Epoch 00370 | Loss 0.3101 | Train Acc. 0.8848 |
Epoch 00375 | Loss 0.3036 | Train Acc. 0.8773 |
Epoch 00380 | Loss 0.2995 | Train Acc. 0.8829 |
Epoch 00385 | Loss 0.3024 | Train Acc. 0.8848 |
Epoch 00390 | Loss 0.2995 | Train Acc. 0.8848 |
Epoch 00395 | Loss 0.2863 | Train Acc. 0.8922 |
Epoch 00400 | Loss 0.2827 | Train Acc. 0.9052 |
Epoch 00405 | Loss 0.2953 | Train Acc. 0.8773 |
Epoch 00410 | Loss 0.2914 | Train Acc. 0.8773 |
Epoch 00415 | Loss 0.2812 | Train Acc. 0.8959 |
Epoch 00420 | Loss 0.2828 | Train Acc. 0.8829 |
Early stopping! No improvement for 100 consecutive epochs.
Fold : 2 | Test Accuracy = 0.8290 | F1 = 0.7999
Total = 42.4Gb Reserved = 0.8Gb Allocated = 0.4Gb
Clearing gpu memory
Total = 42.4Gb Reserved = 0.3Gb Allocated = 0.3Gb
<Figure size 640x480 with 0 Axes>
accuracy = []
F1 = []
i = 0
for metric in output_metrics :
accuracy.append(metric[1])
F1.append(metric[2])
print("%i Fold Cross Validation Accuracy = %2.2f \u00B1 %2.2f" %(5 , np.mean(accuracy)*100 , np.std(accuracy)*100))
print("%i Fold Cross Validation F1 = %2.2f \u00B1 %2.2f" %(5 , np.mean(F1)*100 , np.std(F1)*100))
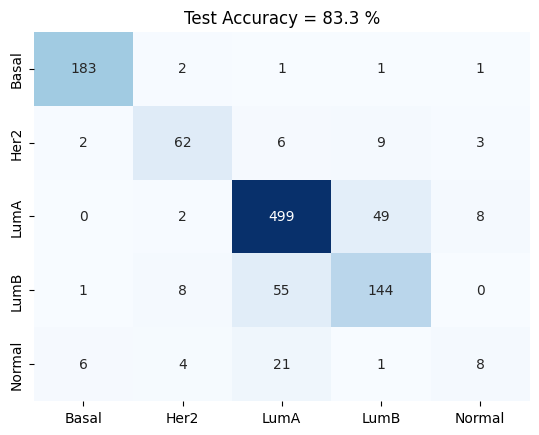
confusion_matrix(test_logits , test_labels , meta.astype('category').cat.categories)
plt.title('Test Accuracy = %2.1f %%' % (np.mean(accuracy)*100))
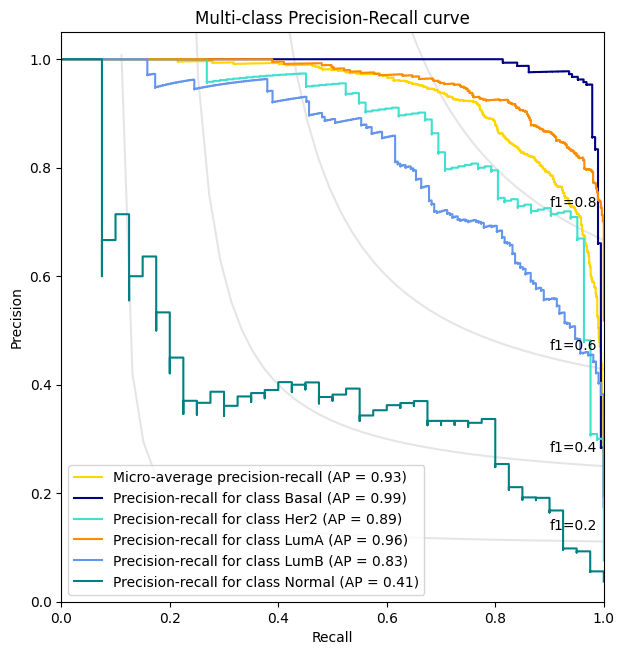
precision_recall_plot , all_predictions_conf = AUROC(test_logits, test_labels , meta)
node_predictions = []
true_label = []
display_label = meta.astype('category').cat.categories
for pred , true in zip(all_predictions_conf.argmax(1) , test_labels.argmax(1).detach().cpu()) :
node_predictions.append(display_label[pred])
true_label.append(display_label[true.item()])
preds = pd.DataFrame({'Actual' : true_label , 'Predicted' : node_predictions})
5 Fold Cross Validation Accuracy = 83.27 ± 0.37
5 Fold Cross Validation F1 = 80.19 ± 0.20