The significance of molecular heterogeneity in breast cancer batch correction and dataset integration
Nicholas Moir, Dominic Pearce, Simon Langdon, T. Ian Simpson
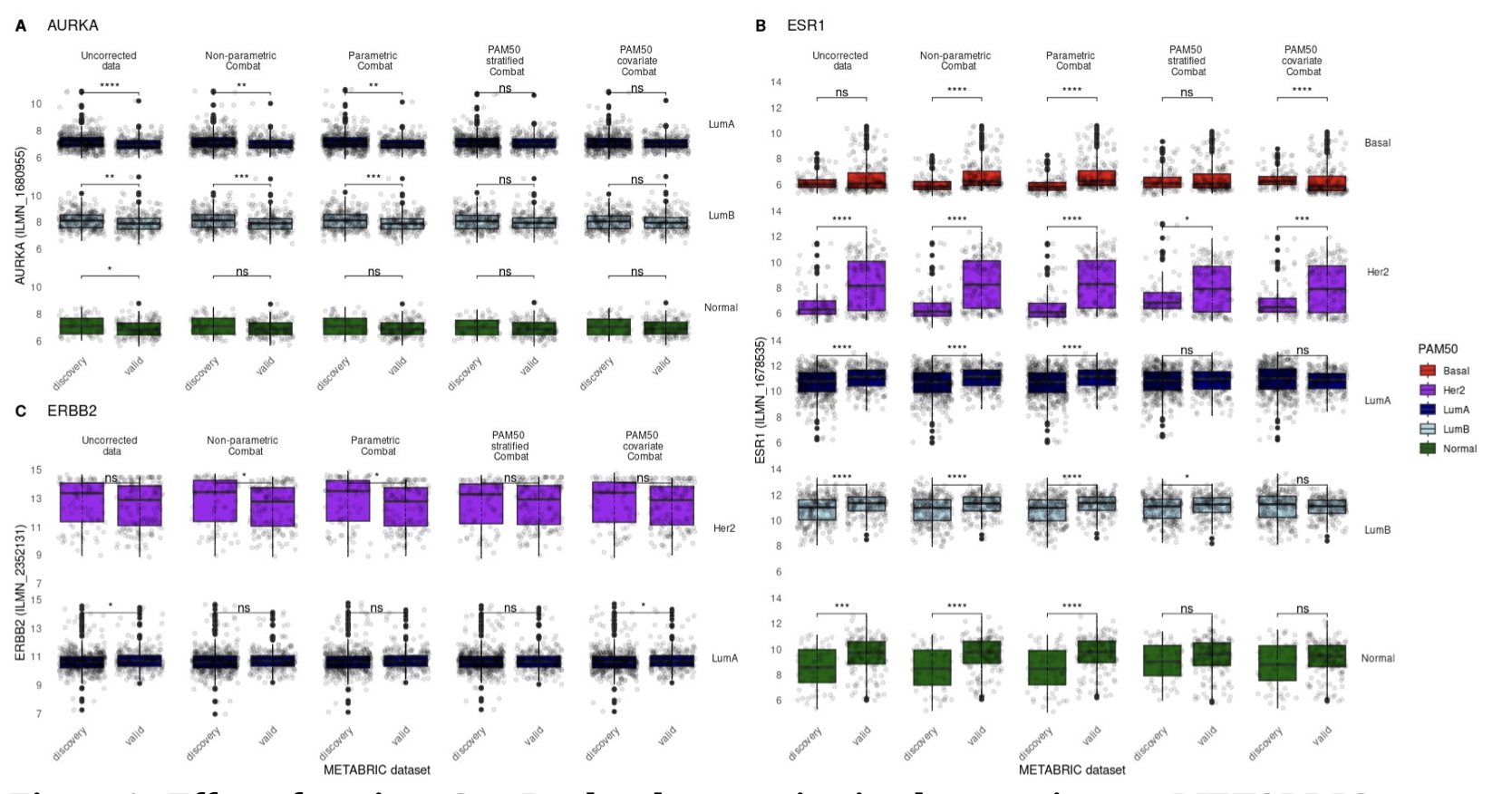
Breast cancer research benefits from a substantial collection of gene expression datasets that are commonly integrated to increase analytical power. Gene expression batch effects arising between experimental batches, where signal differences confound true biological variation, must be addressed when integrating datasets and several approaches exist to address these technical differences. In this work we demonstrate that popular batch correction techniques can significantly distort key biomarker expression signals. Through the implementation of ComBat batch correction and evaluation of integrated expression values, we profile the extent of these distortions and consider an additional mitigatory batch correction step.
We demonstrate that leveraging a priori knowledge of sample molecular subtype classification can optimally remove batch effect distortion while preserving key biomarker expression variation and transcriptional legitimacy. To the best of our knowledge, this study presents the first analysis of the interplay between dataset molecular composition and the concomitant robustness of integrated, batch-corrected biological expression signal.
For more information find the preprint to our paper online here