Combining Clinical Embeddings with Multi-Omic Features for Improved Patient Classification and Interpretability in Parkinsons Disease
Barry Ryan, Chaeeun Lee, Riccardo Marioni, Pasquale Minervini, T. Ian Simpson
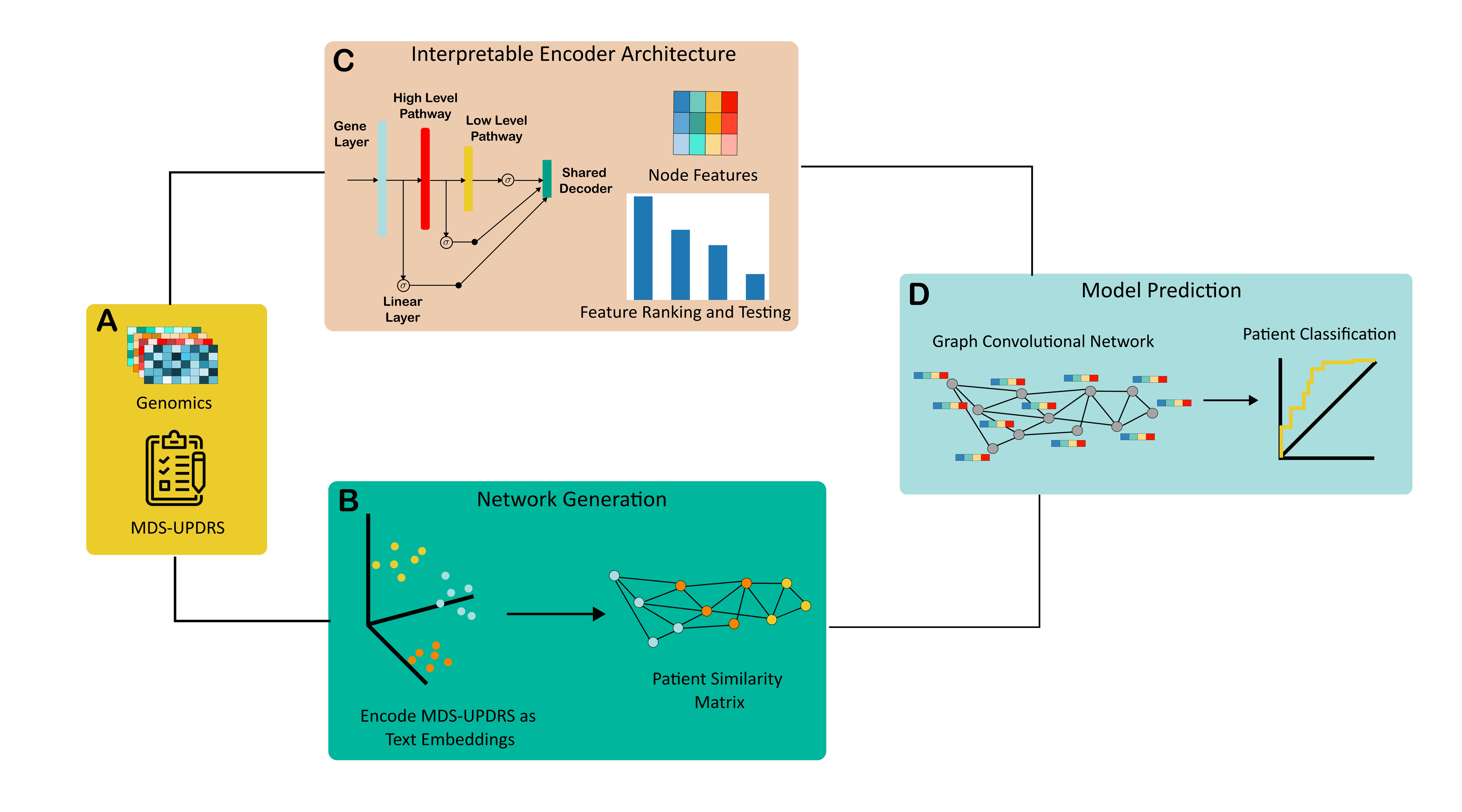
In this work we demonstrate how integration of Large Language Model (LLM)-derived clinical text embeddings from the MDS-UPDRS questionnaire with molecular genomics data can enhance patient classification and interpretability in Parkinsons Disease. By combining genomic modalities encoded using an interpretable biological architecture with a patient similarity network constructed from clinical text embeddings, we leverage clinical and genomic information to provide a robust, interpretable model for disease classification and molecular insights. This work demonstrates that the combination of clinical text embeddings with genomic features is critical for classification and interpretation.
LLM text embeddings not only increase classification accuracy but also enable interpretable genomic analysis, revealing molecular signatures associated with PD progression. Using this framework, we were able to replicate the association of MAPK in PD in a heterogenous cohort from the Parkinsons Progression Markers Initiative.
For more information find the preprint to our paper online here